class: center, middle, inverse, title-slide # Part 5 ## R Programming ### Jessica Minnier ### 2022-02-02 --- class: center, middle # Saving objects --- # Penguins data ```r library(tidyverse) library(janitor) library(palmerpenguins) data(penguins) glimpse(penguins) ``` ``` ## Rows: 344 ## Columns: 8 ## $ species <fct> Adelie, Adelie, Adelie, Adelie,… ## $ island <fct> Torgersen, Torgersen, Torgersen… ## $ bill_length_mm <dbl> 39.1, 39.5, 40.3, NA, 36.7, 39.… ## $ bill_depth_mm <dbl> 18.7, 17.4, 18.0, NA, 19.3, 20.… ## $ flipper_length_mm <int> 181, 186, 195, NA, 193, 190, 18… ## $ body_mass_g <int> 3750, 3800, 3250, NA, 3450, 365… ## $ sex <fct> male, female, female, NA, femal… ## $ year <int> 2007, 2007, 2007, 2007, 2007, 2… ``` --- # When we filter/clean/mutate the data - If you're changing the data and you want to use that data later, save your work - Save as new object, *or*, - Save as the same object if you want to overwrite that data frame within R ```r # save as new data frame penguins_f penguins_f <- penguins %>% filter(sex=="female") %>% mutate(bill_ratio = bill_length_mm/bill_depth_mm) # change the ordering of penguins_f and re-save it penguins_f <- penguins_f %>% arrange(bill_ratio) # print out that data frame, but don't save it penguins_f %>% select(sex, contains("bill")) ``` ``` ## # A tibble: 165 × 4 ## sex bill_length_mm bill_depth_mm bill_ratio ## <fct> <dbl> <dbl> <dbl> ## 1 female 33.5 19 1.76 ## 2 female 35.3 18.9 1.87 ## 3 female 34.4 18.4 1.87 ## 4 female 35.9 19.2 1.87 ## 5 female 36.7 19.3 1.90 ## 6 female 34.5 18.1 1.91 ## 7 female 39.6 20.7 1.91 ## 8 female 36 18.5 1.95 ## 9 female 37.6 19.3 1.95 ## 10 female 36.7 18.8 1.95 ## # … with 155 more rows ``` --- # Summarizing data (i.e. tabyl, summarize) - `tabyl()` and `summarize()` create new tibbles/data.frames that have summary info - You can save it if you want, but not usually necessary unless you want to use it later! - From `?tabyl`, Value = "Returns a data.frame with frequencies and percentages of the tabulated variable(s). A 3-way tabulation returns a list of data.frames." .pull-left-40[ ```r penguins_f %>% tabyl(species) ``` ``` ## species n percent ## Adelie 73 0.4424242 ## Chinstrap 34 0.2060606 ## Gentoo 58 0.3515152 ``` ] .pull-right-60[ ```r # save output as a tibble penguins_f_table <- penguins_f %>% tabyl(species) # we can see this is a tibble! glimpse(penguins_f_table) ``` ``` ## Rows: 3 ## Columns: 3 ## $ species <fct> Adelie, Chinstrap, Gentoo ## $ n <dbl> 73, 34, 58 ## $ percent <dbl> 0.4424242, 0.2060606, 0.3515152 ``` ```r class(penguins_f_table) ``` ``` ## [1] "tabyl" "data.frame" ``` ] --- # Using summaries later ```r penguins_f_table %>% arrange(n) ``` ``` ## species n percent ## Chinstrap 34 0.2060606 ## Gentoo 58 0.3515152 ## Adelie 73 0.4424242 ``` Nice printing: ```r penguins_f_table %>% gt::gt() ``` <div id="fwwjzjwxfh" style="overflow-x:auto;overflow-y:auto;width:auto;height:auto;"> <style>html { font-family: -apple-system, BlinkMacSystemFont, 'Segoe UI', Roboto, Oxygen, Ubuntu, Cantarell, 'Helvetica Neue', 'Fira Sans', 'Droid Sans', Arial, sans-serif; } #fwwjzjwxfh .gt_table { display: table; border-collapse: collapse; margin-left: auto; margin-right: auto; color: #333333; font-size: 16px; font-weight: normal; font-style: normal; background-color: #FFFFFF; width: auto; border-top-style: solid; border-top-width: 2px; border-top-color: #A8A8A8; border-right-style: none; border-right-width: 2px; border-right-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #A8A8A8; border-left-style: none; border-left-width: 2px; border-left-color: #D3D3D3; } #fwwjzjwxfh .gt_heading { background-color: #FFFFFF; text-align: center; border-bottom-color: #FFFFFF; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; } #fwwjzjwxfh .gt_title { color: #333333; font-size: 125%; font-weight: initial; padding-top: 4px; padding-bottom: 4px; border-bottom-color: #FFFFFF; border-bottom-width: 0; } #fwwjzjwxfh .gt_subtitle { color: #333333; font-size: 85%; font-weight: initial; padding-top: 0; padding-bottom: 6px; border-top-color: #FFFFFF; border-top-width: 0; } #fwwjzjwxfh .gt_bottom_border { border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; } #fwwjzjwxfh .gt_col_headings { border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; } #fwwjzjwxfh .gt_col_heading { color: #333333; background-color: #FFFFFF; font-size: 100%; font-weight: normal; text-transform: inherit; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; vertical-align: bottom; padding-top: 5px; padding-bottom: 6px; padding-left: 5px; padding-right: 5px; overflow-x: hidden; } #fwwjzjwxfh .gt_column_spanner_outer { color: #333333; background-color: #FFFFFF; font-size: 100%; font-weight: normal; text-transform: inherit; padding-top: 0; padding-bottom: 0; padding-left: 4px; padding-right: 4px; } #fwwjzjwxfh .gt_column_spanner_outer:first-child { padding-left: 0; } #fwwjzjwxfh .gt_column_spanner_outer:last-child { padding-right: 0; } #fwwjzjwxfh .gt_column_spanner { border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; vertical-align: bottom; padding-top: 5px; padding-bottom: 5px; overflow-x: hidden; display: inline-block; width: 100%; } #fwwjzjwxfh .gt_group_heading { padding: 8px; color: #333333; background-color: #FFFFFF; font-size: 100%; font-weight: initial; text-transform: inherit; border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; vertical-align: middle; } #fwwjzjwxfh .gt_empty_group_heading { padding: 0.5px; color: #333333; background-color: #FFFFFF; font-size: 100%; font-weight: initial; border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; vertical-align: middle; } #fwwjzjwxfh .gt_from_md > :first-child { margin-top: 0; } #fwwjzjwxfh .gt_from_md > :last-child { margin-bottom: 0; } #fwwjzjwxfh .gt_row { padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; margin: 10px; border-top-style: solid; border-top-width: 1px; border-top-color: #D3D3D3; border-left-style: none; border-left-width: 1px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 1px; border-right-color: #D3D3D3; vertical-align: middle; overflow-x: hidden; } #fwwjzjwxfh .gt_stub { color: #333333; background-color: #FFFFFF; font-size: 100%; font-weight: initial; text-transform: inherit; border-right-style: solid; border-right-width: 2px; border-right-color: #D3D3D3; padding-left: 12px; } #fwwjzjwxfh .gt_summary_row { color: #333333; background-color: #FFFFFF; text-transform: inherit; padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; } #fwwjzjwxfh .gt_first_summary_row { padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; } #fwwjzjwxfh .gt_grand_summary_row { color: #333333; background-color: #FFFFFF; text-transform: inherit; padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; } #fwwjzjwxfh .gt_first_grand_summary_row { padding-top: 8px; padding-bottom: 8px; padding-left: 5px; padding-right: 5px; border-top-style: double; border-top-width: 6px; border-top-color: #D3D3D3; } #fwwjzjwxfh .gt_striped { background-color: rgba(128, 128, 128, 0.05); } #fwwjzjwxfh .gt_table_body { border-top-style: solid; border-top-width: 2px; border-top-color: #D3D3D3; border-bottom-style: solid; border-bottom-width: 2px; border-bottom-color: #D3D3D3; } #fwwjzjwxfh .gt_footnotes { color: #333333; background-color: #FFFFFF; border-bottom-style: none; border-bottom-width: 2px; border-bottom-color: #D3D3D3; border-left-style: none; border-left-width: 2px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 2px; border-right-color: #D3D3D3; } #fwwjzjwxfh .gt_footnote { margin: 0px; font-size: 90%; padding: 4px; } #fwwjzjwxfh .gt_sourcenotes { color: #333333; background-color: #FFFFFF; border-bottom-style: none; border-bottom-width: 2px; border-bottom-color: #D3D3D3; border-left-style: none; border-left-width: 2px; border-left-color: #D3D3D3; border-right-style: none; border-right-width: 2px; border-right-color: #D3D3D3; } #fwwjzjwxfh .gt_sourcenote { font-size: 90%; padding: 4px; } #fwwjzjwxfh .gt_left { text-align: left; } #fwwjzjwxfh .gt_center { text-align: center; } #fwwjzjwxfh .gt_right { text-align: right; font-variant-numeric: tabular-nums; } #fwwjzjwxfh .gt_font_normal { font-weight: normal; } #fwwjzjwxfh .gt_font_bold { font-weight: bold; } #fwwjzjwxfh .gt_font_italic { font-style: italic; } #fwwjzjwxfh .gt_super { font-size: 65%; } #fwwjzjwxfh .gt_footnote_marks { font-style: italic; font-weight: normal; font-size: 65%; } </style> <table class="gt_table"> <thead class="gt_col_headings"> <tr> <th class="gt_col_heading gt_columns_bottom_border gt_center" rowspan="1" colspan="1">species</th> <th class="gt_col_heading gt_columns_bottom_border gt_right" rowspan="1" colspan="1">n</th> <th class="gt_col_heading gt_columns_bottom_border gt_right" rowspan="1" colspan="1">percent</th> </tr> </thead> <tbody class="gt_table_body"> <tr><td class="gt_row gt_center">Adelie</td> <td class="gt_row gt_right">73</td> <td class="gt_row gt_right">0.4424242</td></tr> <tr><td class="gt_row gt_center">Chinstrap</td> <td class="gt_row gt_right">34</td> <td class="gt_row gt_right">0.2060606</td></tr> <tr><td class="gt_row gt_center">Gentoo</td> <td class="gt_row gt_right">58</td> <td class="gt_row gt_right">0.3515152</td></tr> </tbody> </table> </div> --- # Using summaries later You can also save your summaries to excel or csv files: ```r # one long pipe penguins_f_table %>% arrange(n) %>% write_csv(file = "table_of_species_in_females.csv") # or save as tibble object and then write tibble to csv file penguins_f_table <- penguins_f_table %>% arrange(n) write_csv(penguins_f_table, file = "table_of_species_in_females.csv") ``` --- # "Printing" or "viewing" Some things you *__don't__* usually want to save - `glimpse()` prints a snapshot of your data, not useful to save - `head()` or `tail()`, there's almost no reason to save just the first few lines of your data, except for educational purposes! - Similarly, you can pipe on `nrow()` or `dim()` or `colnames()` or `class()` and so on, these are pulling out meta-data/information from your data; you can save this as an object but make sure you know what the output is! (probably a vector of information) - `skim()` prints a visual summary of your data, you can save it if you want to do fancy things with it - `view()`/`View()` pops up a window that shows your data, don't save this and don't keep this code in your Rmd --- ### glimpse ```r # when you save this, it prints it, and then you could print it again glimpse_out <- glimpse(penguins_f) ``` ``` ## Rows: 165 ## Columns: 9 ## $ species <fct> Adelie, Adelie, Adelie, Adelie,… ## $ island <fct> Torgersen, Biscoe, Torgersen, B… ## $ bill_length_mm <dbl> 33.5, 35.3, 34.4, 35.9, 36.7, 3… ## $ bill_depth_mm <dbl> 19.0, 18.9, 18.4, 19.2, 19.3, 1… ## $ flipper_length_mm <int> 190, 187, 184, 189, 193, 187, 1… ## $ body_mass_g <int> 3600, 3800, 3325, 3800, 3450, 2… ## $ sex <fct> female, female, female, female,… ## $ year <int> 2008, 2007, 2007, 2007, 2007, 2… ## $ bill_ratio <dbl> 1.763158, 1.867725, 1.869565, 1… ``` Print it now (but really, don't do this, there's no point): ```r glimpse_out ``` ``` ## # A tibble: 165 × 9 ## species island bill_length_mm bill_depth_mm ## <fct> <fct> <dbl> <dbl> ## 1 Adelie Torgersen 33.5 19 ## 2 Adelie Biscoe 35.3 18.9 ## 3 Adelie Torgersen 34.4 18.4 ## 4 Adelie Biscoe 35.9 19.2 ## 5 Adelie Torgersen 36.7 19.3 ## 6 Adelie Biscoe 34.5 18.1 ## 7 Adelie Biscoe 39.6 20.7 ## 8 Adelie Dream 36 18.5 ## 9 Adelie Dream 37.6 19.3 ## 10 Adelie Torgersen 36.7 18.8 ## # … with 155 more rows, and 5 more variables: ## # flipper_length_mm <int>, body_mass_g <int>, sex <fct>, ## # year <int>, bill_ratio <dbl> ``` --- ### skim from `?skim`: Value = "A skim_df object, which also inherits the class(es) of the input data. In many ways, the object behaves like a `tibble::tibble()`." ```r # when you save this, it doesn't print it, but you can print it later skim_out <- skimr::skim(penguins_f) ``` Now you can print it (this gets cut off on the slide) ```r skim_out ``` Table: Data summary | | | |:------------------------|:----------| |Name |penguins_f | |Number of rows |165 | |Number of columns |9 | |_______________________ | | |Column type frequency: | | |factor |3 | |numeric |6 | |________________________ | | |Group variables |None | **Variable type: factor** |skim_variable | n_missing| complete_rate|ordered | n_unique|top_counts | |:-------------|---------:|-------------:|:-------|--------:|:-------------------------| |species | 0| 1|FALSE | 3|Ade: 73, Gen: 58, Chi: 34 | |island | 0| 1|FALSE | 3|Bis: 80, Dre: 61, Tor: 24 | |sex | 0| 1|FALSE | 1|fem: 165, mal: 0 | **Variable type: numeric** |skim_variable | n_missing| complete_rate| mean| sd| p0| p25| p50| p75| p100|hist | |:-----------------|---------:|-------------:|-------:|------:|-------:|-------:|-------:|-------:|-------:|:-----| |bill_length_mm | 0| 1| 42.10| 4.90| 32.10| 37.60| 42.80| 46.20| 58.00|▅▅▇▂▁ | |bill_depth_mm | 0| 1| 16.43| 1.80| 13.10| 14.50| 17.00| 17.80| 20.70|▇▃▇▇▁ | |flipper_length_mm | 0| 1| 197.36| 12.50| 172.00| 187.00| 193.00| 210.00| 222.00|▂▇▃▃▃ | |body_mass_g | 0| 1| 3862.27| 666.17| 2700.00| 3350.00| 3650.00| 4550.00| 5200.00|▃▇▂▃▃ | |year | 0| 1| 2008.04| 0.81| 2007.00| 2007.00| 2008.00| 2009.00| 2009.00|▇▁▇▁▇ | |bill_ratio | 0| 1| 2.61| 0.50| 1.76| 2.16| 2.54| 3.12| 3.49|▇▇▅▆▇ | --- You can `glimpse()` the skim output because it's deep down a tibble! ```r class(skim_out) ``` ``` ## [1] "skim_df" "tbl_df" "tbl" "data.frame" ``` ```r glimpse(skim_out) ``` ``` ## Rows: 9 ## Columns: 15 ## $ skim_type <chr> "factor", "factor", "factor", "… ## $ skim_variable <chr> "species", "island", "sex", "bi… ## $ n_missing <int> 0, 0, 0, 0, 0, 0, 0, 0, 0 ## $ complete_rate <dbl> 1, 1, 1, 1, 1, 1, 1, 1, 1 ## $ factor.ordered <lgl> FALSE, FALSE, FALSE, NA, NA, NA… ## $ factor.n_unique <int> 3, 3, 1, NA, NA, NA, NA, NA, NA ## $ factor.top_counts <chr> "Ade: 73, Gen: 58, Chi: 34", "B… ## $ numeric.mean <dbl> NA, NA, NA, 42.096970, 16.42545… ## $ numeric.sd <dbl> NA, NA, NA, 4.9034759, 1.795680… ## $ numeric.p0 <dbl> NA, NA, NA, 32.100000, 13.10000… ## $ numeric.p25 <dbl> NA, NA, NA, 37.600000, 14.50000… ## $ numeric.p50 <dbl> NA, NA, NA, 42.800000, 17.00000… ## $ numeric.p75 <dbl> NA, NA, NA, 46.200000, 17.80000… ## $ numeric.p100 <dbl> NA, NA, NA, 58.000000, 20.70000… ## $ numeric.hist <chr> NA, NA, NA, "▅▅▇▂▁", "▇▃▇▇▁", "… ``` --- See `?skim` for examples on what you can do with `skim()` output, as well as the [skimr vignette](https://cran.r-project.org/web/packages/skimr/vignettes/skimr.html) ```r skim_out %>% filter(n_missing > 0) ``` ``` ## # A tibble: 0 × 15 ## # … with 15 variables: skim_type <chr>, ## # skim_variable <chr>, n_missing <int>, ## # complete_rate <dbl>, factor.ordered <lgl>, ## # factor.n_unique <int>, factor.top_counts <chr>, ## # numeric.mean <dbl>, numeric.sd <dbl>, ## # numeric.p0 <dbl>, numeric.p25 <dbl>, ## # numeric.p50 <dbl>, numeric.p75 <dbl>, … ``` ```r skim_out %>% filter(skim_type == "numeric") %>% select(skim_variable, numeric.mean) ``` ``` ## # A tibble: 6 × 2 ## skim_variable numeric.mean ## <chr> <dbl> ## 1 bill_length_mm 42.1 ## 2 bill_depth_mm 16.4 ## 3 flipper_length_mm 197. ## 4 body_mass_g 3862. ## 5 year 2008. ## 6 bill_ratio 2.61 ``` --- ### view - Use `View` or `view()` interactively, don't include this code in your Rmd, and don't save it as an object! ```r View(penguins_f) # this will pop up every time you knit, not necessary pview <- penguins_f %>% View() # this is not helpful ``` pview is `NULL` because `View()` produces NO output (no value) ```r pview ``` ``` ## NULL ``` --- # Saving ggplot - Save a ggplot *as an object* if you want to "call it" later to add more layers, or if you want to save it *as an image file* (i.e .png, .pdf, .tif, etc) on your computer - `ggsave()` by default saves the last plot printed to a file, but to be extra careful you can use the object name ```r # save as object p_boxplot; will not print out plot p_boxplot <- ggplot(penguins, aes(x = species, y = bill_length_mm, fill = sex)) + geom_boxplot() # it's a ggplot object class(p_boxplot) ``` ``` ## [1] "gg" "ggplot" ``` ```r # print the plot in output p_boxplot ``` ``` ## Warning: Removed 2 rows containing non-finite values ## (stat_boxplot). ``` 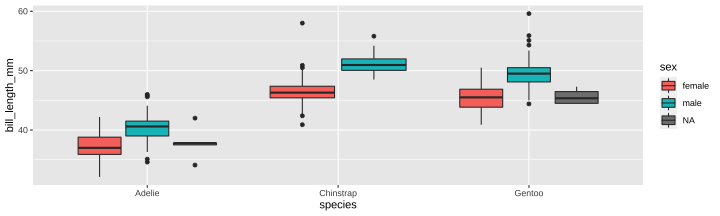<!-- --> --- Now we can add additional layers, and/or save it as a png file! ```r # save as png (default location is same folder where Rmd is) ggsave(p_boxplot, file = "p_boxplot.png", height = 6, width = 5) ``` ``` ## Warning: Removed 2 rows containing non-finite values ## (stat_boxplot). ``` ```r # add layers p_boxplot + theme_minimal() + labs(title = "Add layers to my boxplot!", x = "") ``` ``` ## Warning: Removed 2 rows containing non-finite values ## (stat_boxplot). ``` 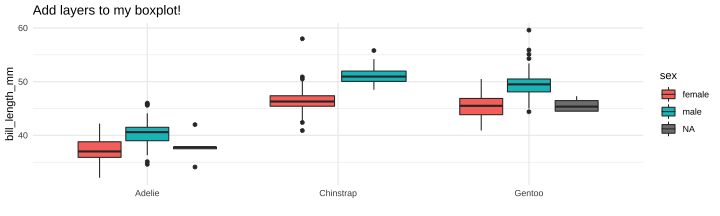<!-- --> --- Want to save that plot? ```r p_boxplot <- p_boxplot + theme_minimal() + labs(title = "Add layers to my boxplot!", x = "") ggsave(p_boxplot, file = "p_boxplot_minimal.png", height = 6, width = 5) ``` ``` ## Warning: Removed 2 rows containing non-finite values ## (stat_boxplot). ``` ```r p_boxplot ``` ``` ## Warning: Removed 2 rows containing non-finite values ## (stat_boxplot). ``` 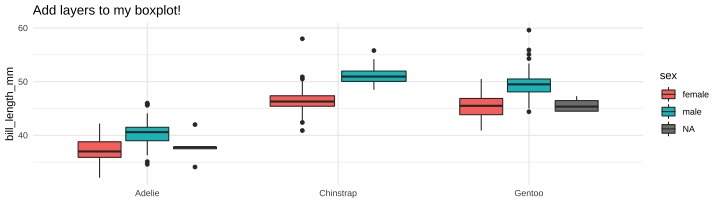<!-- --> --- # Objects - All of this has to do with classes and objects in R, which is an object-oriented programming language. - `class(objectname)` tells us what kind of structure it is and how we can use it - Many functions output a unique kind of object ```r tmpfit <- lm(bill_length_mm ~ species, data = penguins) class(tmpfit) ``` ``` ## [1] "lm" ``` ```r # see Value section of ?lm ``` - If you want some bedtime reading on this, start with the [Advanced R textbook's "OO field guide"](http://adv-r.had.co.nz/OO-essentials.html)